Upload X-ray Dataset
This guide provides instructions for uploading X-ray crystallography data to CryoCloud for automated structure refinement and analysis.
Overview
X-ray datasets in CryoCloud typically consist of diffraction experiments from a single synchrotron trip, provided either as raw images or pre-processed datasets. Raw data is identified by *_master.h5 files or a sufficient number of cbf files with a common file prefix. For pre-processing software, we support metadata extraction from autoPROC, xia2-* (variations of DIALS and 3D), and Grenades. Please feel free to reach out to us if you want to upload other pre-processed mtz files.
Our platform provides access to the automated pipedream pipeline by Global Phasing, which can refine structures and generate high-resolution models with minimal manual intervention.
Preparing Your Data
Before uploading, ensure your X-ray dataset includes:
- Diffraction data - Raw or pre-processed diffraction data files from your beamline
- Pre-processing metadata files (optional) - Any associated metadata files
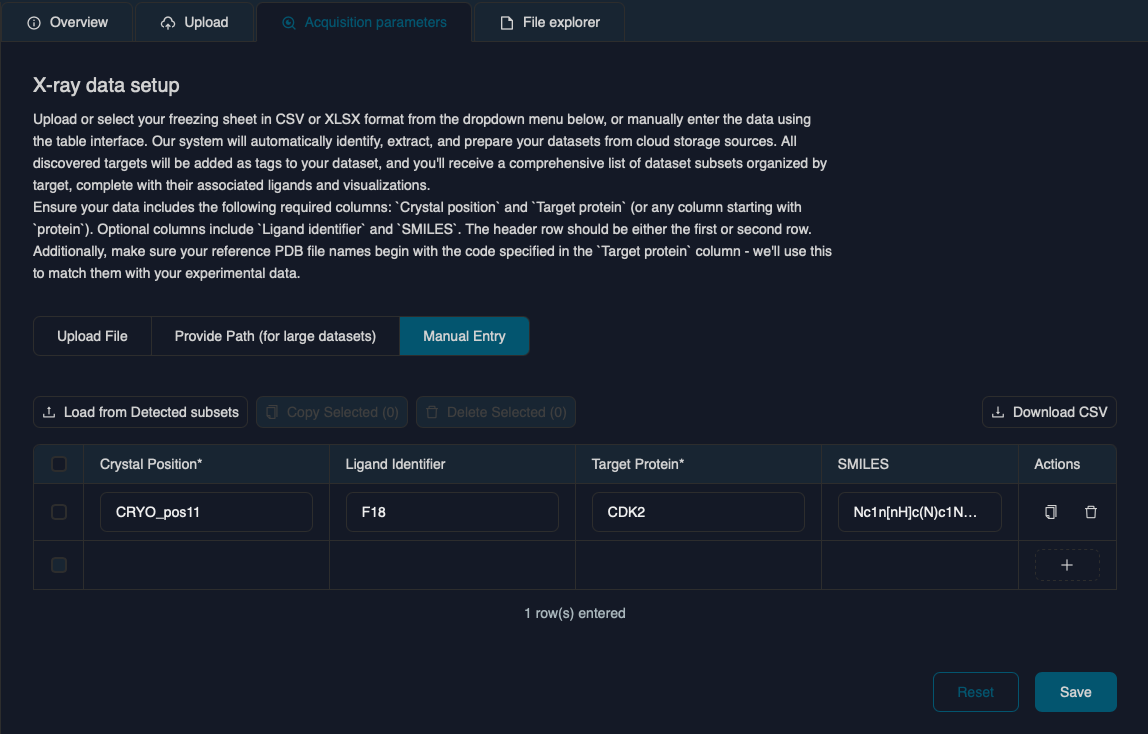
- Synchrotron freezing sheet - A spreadsheet file with the description of the collected data in either CSV or XLSX format. Please find the minimum required CSV file template here
- Reference structures - Reference coordinates in PDB format
Dataset Structure and Upload
General guidelines for dataset upload can be found on the Dataset page. Here we focus on specifics associated with X-ray data.
Data Organization
The dataset scanning method follows these steps:
- Parse the metadata spreadsheet containing Crystal Position, Target Protein, and optional Ligand Information (ID and SMILES string). Alternatively, you can supply this information via manual input
- Identify data subfolders in cloud storage (raw images and pre-processed data) based on common filenames
- Match Crystal Positions to data using path matching logic:
<puckID>_<crystal_position>is searched in the identified file paths (or directory paths in the case of raw data) - Extract metadata from pre-processing logs and unit cell parameters from reference PDB files:
- We look for any PDB files in the bucket starting with the Protein Acronym/Target found in the CSV/XLSX
- Create dataset entries by pairing ligands with their experimental data
After ensuring your uploaded data follows these guidelines, click Save to proceed. After a few moments, you should see your dataset populated.
Next Steps
Once your X-ray dataset is uploaded and configured, you can create a Workflow in the Workflows tab. Currently, it is only necessary to add a single Node of XRayRefine Job to a workflow, adjust the desired parameters, and then navigate to the Projects tab to start a Project from the Workflow you just created.
Troubleshooting
Upload Issues:
If an expected subset is missing:
- Ensure the spreadsheet specifies both the Crystal Position and the Protein Target.
- Upload at least one reference PDB model for each Protein Target.
- Confirm the software used is supported (metadata is extracted from log files).
- Check for duplicate crystal position records in your spreadsheet.
Need Help?
If you encounter issues with X-ray dataset upload or have questions about data formats:
- Contact CryoCloud support
- Refer to our X-ray Tutorial for a complete workflow example
- Check our Support page for additional resources